Beginning Data Science with Jupyter Notebook and Kotlin
This tutorial introduces the concepts of Data Science, using Jupyter Notebook and Kotlin. You’ll learn how to set up a Jupyter notebook, load krangl for Kotlin and use it in data science utilizing a built-in sample data. By Joey deVilla.
Sign up/Sign in
With a free Kodeco account you can download source code, track your progress, bookmark, personalise your learner profile and more!
Create accountAlready a member of Kodeco? Sign in
Sign up/Sign in
With a free Kodeco account you can download source code, track your progress, bookmark, personalise your learner profile and more!
Create accountAlready a member of Kodeco? Sign in
Sign up/Sign in
With a free Kodeco account you can download source code, track your progress, bookmark, personalise your learner profile and more!
Create accountAlready a member of Kodeco? Sign in
Contents
Beginning Data Science with Jupyter Notebook and Kotlin
35 mins
- Getting Started
- Working with krangl’s Built-In Data Frames
- Getting a Data Frame’s First and Last Rows
- Extracting a slice() from the Data Frame
- Learning that slice() Indexes Start at 1, not 0
- Exploring the Data
- Sorting Data
- Filtering
- Learning the Basics of Filtering
- Filtering With Negation
- Filtering With Multiple Criteria
- Fancier Text Filtering
- Removing Columns
- Performing Complex Data Operations
- Calculating Column Statistics
- Grouping
- Summarizing
- Importing Data
- Reading .csv Data
- Reading .tsv Data
- Where to Go From Here?
This article builds on Create Your Own Kotlin Playground (and Get a Data Science Head Start) with Jupyter Notebook, which covers the following:
- Jupyter Notebook, an open source web application that lets you create documents that mix text, graphics, and live code. Its native programming language is Python.
- The Kotlin kernel for Jupyter Notebook adds a Kotlin interpreter. The previous article showed how you could use Jupyter Notebook and the Kotlin Kernel as an interactive “playground” for Kotlin code.
- krangl, a library whose name is short for Kotlin library for data wrangling.
- Data frames, which are data structures that represent tables of data organized into rows and columns. They’re the primary data structure in data science applications.
In this article, you’ll take what you’ve learned and build on it by performing basic data science.
Getting Started
If you’re not already running Jupyter Notebook, launch it now. The simplest way to do so is to enter the following on the command line:
jupyter notebook
Your computer will open a new browser window or tab, and it will contain a page that looks similar to this:

Create a new Kotlin notebook by clicking on the New button near the page’s upper right corner. A menu will appear, one of the options will be Kotlin; select that option to create a new Kotlin notebook.
A new browser tab or window will open, containing a page that looks like this:

The page will contain a single cell, a code cell (the default). Enter the following into the cell:
%use krangl
Now run it, either by clicking the Run button or by typing Shift-Enter. This will import krangl.
The cell should look like this for a few seconds…

…then it will look like this:

At this point, krangl should have loaded, and you can now use it for data science.
Working with krangl’s Built-In Data Frames
People who are just getting started with data science often struggle with finding good datasets to work with. To counter this problem, krangl provides three datasets in the form of three built-in DataFrame instances.
Going from smallest to largest, these built-in data frames are:
-
sleepData: Data about the sleep patterns of 83 species of animal, including humans. -
irisData: A set of measurements of 150 flowers from different species of iris. -
flightsData: Over 330,000 rows of data about flights taking off from New York City airports in 2013.
Getting a Data Frame’s First and Last Rows
The datasets that you’ll often encounter will contain a lot of observations, possibly hundreds, thousands, or even millions. In order not to overwhelm you, krangl limits the amount it displays by default.
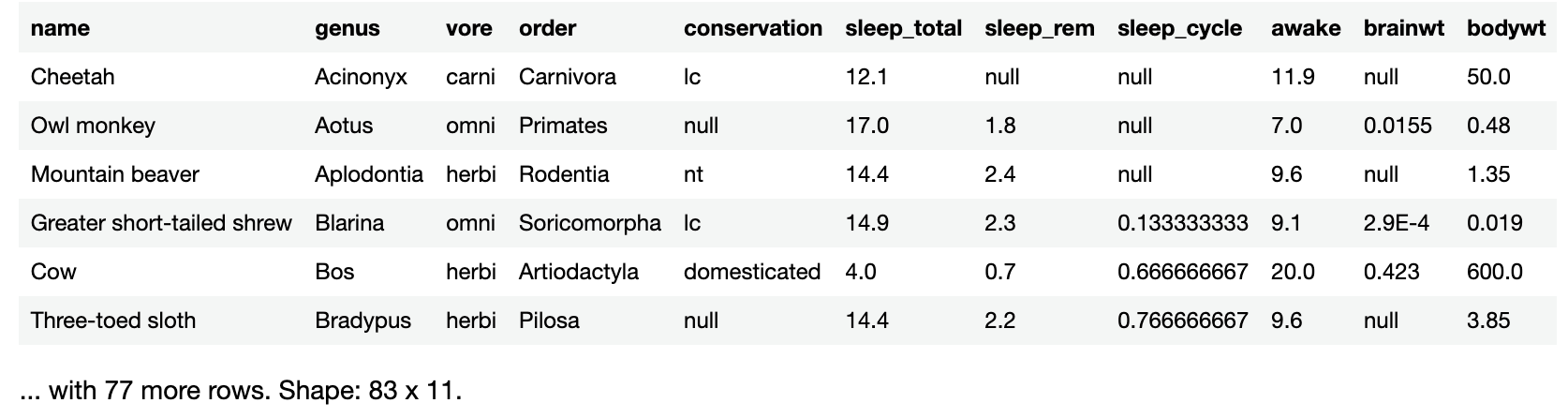
If you simply enter the name of a large data frame into a code cell and run it, you’ll see only the first six rows, followed by a count of the remaining rows and the data frame’s dimensions.
Try it with sleepData. Run the following in a new code cell:
sleepData
You’ll see this result:
Notice that krangl tells you how many more rows there are in the data frame, 77 in this case, as well as the data frame’s dimensions: 83 rows and 11 columns. This is important because it tells you that while krangl is only showing you the first 6 rows, sleepData actually has 83 rows of data.
To see only the first n rows of the data frame, you can use the head() method. Run the following in a new code cell:
sleepData.head()
The notebook will respond by returning a data frame made up of the first five rows of sleepData:
head() returns a new data frame made from the first n rows of the original data frame. By default, n is 5.
Try getting the first 20 rows of sleepData by running the following in a new code cell:
sleepData.head(20)
You’ll see these results:
The result is a data frame consisting of the first 20 rows of sleepData, with the first six rows displayed onscreen. The text at the bottom of the data frame shows you how many rows you’re not seeing, as well as the data frame’s dimensions.
head() has a counterpart, tail(), which returns a new data frame made up of the last n rows of the original data frame. As with head(), the default value for n is 5.
Run the following in a new code cell:
val lastFew = sleepData.tail() lastFew
The notebook will respond by creating and displaying lastFew, a data frame that contains the last 5 rows of sleepData:
Extracting a slice() from the Data Frame
Suppose you weren’t interested in the first or last n rows of the data frame, but a section in the middle. That’s where the slice() method comes in. Given an integer range, it creates a new data frame consisting of the rows specified in the range.
Run the following in a new code cell:
val selection = sleepData.slice(30..34) selection
This creates a new data frame, selection, and then displays its contents:
If you looked at the row for “Human” and thought, “Who actually gets eight hours of sleep?!” keep in mind that these numbers are averages.
Learning that slice() Indexes Start at 1, not 0
Krangl borrows the DataFrame‘s slice()method from the dplyr library.
Dplyr is a library for R, where the first index of a collection type is 1. This is known as one-based array indexing.
In Kotlin, and most other programming languages, the first index of a collection type is 0.
krangl makes it easy for R programmers to make the leap to doing Kotlin sdata science. One of the ways to meet this goal would be to make krangl’s version of slice() work exactly like dplyr’s, right down to using 1 as the index for the first row in the data frame.
It’s time to do a little exploring.
Exploring the Data
Look back at the code cell where you used head() to see the first 5 rows of sleepData:
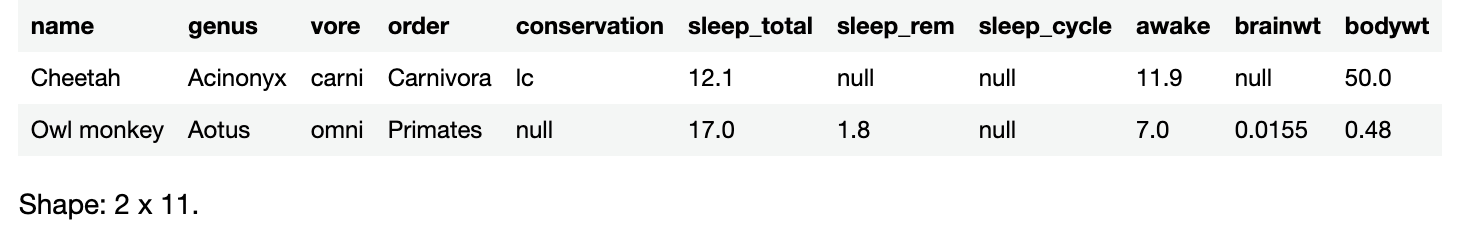
The first animal in the list is the cheetah. Try using DataFrame’s rows property and rows elementAt() method to retrieve the row whose index is 0:
sleepData.rows.elementAt(0)
You should see this result:
{name=Cheetah, genus=Acinonyx, vore=carni, order=Carnivora, conservation=lc, sleep_total=12.1, sleep_rem=null, sleep_cycle=null, awake=11.9, brainwt=null, bodywt=50.0}
This makes sense as rows is a Kotlin Iterable and elementAt() is a method of Iterable. It makes sense that sleepData.rows.elementAt(0) would return an object representing the first row of sleepData.
Now, try approximating the same thing with slice(). The result will be of a different type: sleepData.rows.elementAt() returns a DataFrameRow, while slice() returns a whole DataFrame.
Run the following in a new code cell:
sleepData.slice(0..0)
The result will be an empty data frame; 11 columns, but no rows:
Now, try something else. Run the following in a new code cell:
sleepData.slice(1..1)
This time, you get a data frame with a single row; note the dimensions:
Try one more thing; run the following in a new code cell:
sleepData.slice(1..2)
This results in a data frame with two rows:
From these little bits of code, you can deduce that slice(a..b) returns a data frame that:
- Starts with and includes row a from the original data frame.
- Ends with and includes row b from the original data frame.
- Counts rows starting from 1 rather than starting from 0.